7.4. Data#
7.4.2. Exercises#
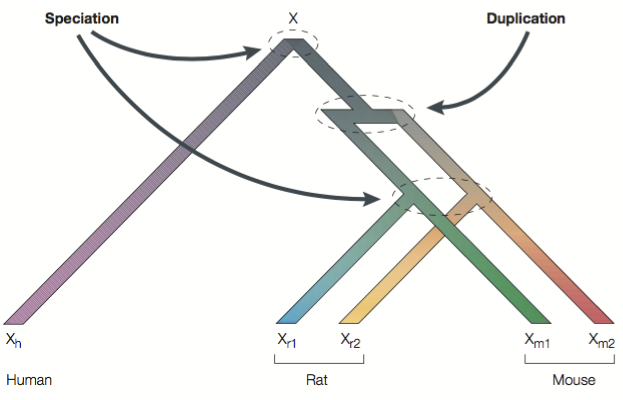
I have queried the Ensembl database for some haemoglobins from Human and Macaque. For each database record, I’ve used Ensembl’s definition of the “canonical transcript” (or “major” transcript). I’ve then obtained the CDS (coding sequence) for each record. I named the sequences {Species Common Name}-{a letter}. I then aligned the sequences.
| 0 | |
| Human-j | ATGGTGCATCTGACTCCTGAGGAGAAGTCT |
| Human-z | ......---...T......CC..C...A.C |
| Rhesus-e | ......---...T......CC..C...AGC |
| Rhesus-x | ...........................AA. |
4 x 447 (truncated to 4 x 30) dna alignment
Which of the above are orthologs?
Which are paralogs?
For these sequences, how would you describe the divergence between orthologs compared to that between paralogs?
Citations
David B. Searls. Pharmacophylogenomics: genes, evolution and drug targets. Nature Reviews Drug Discovery, 2:613–623, 2003. URL: https://doi.org/10.1038/nrd1152, doi:10.1038/nrd1152.